Interactive: Click the work package (WP) for more information!
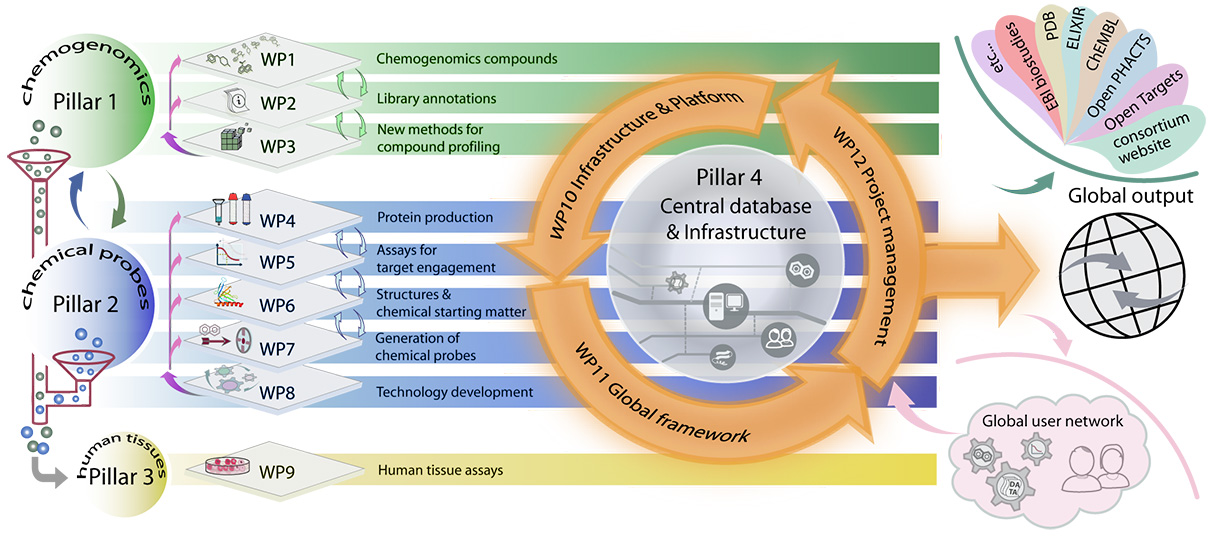
WP1 aims to create a “first generation” Chemogenomics Library (CGL) comprising approximately 2,000 known compounds, which will cover at least 500 targets. Compounds will be acquired in sufficient amounts for distribution and each compound will fulfil stringent quality criteria. This set will be assembled from available chemical probe and chemogenomic sets and augmented with probes from the literature. WP1 interacts with WP2 to annotate compounds and their properties leading to a CGL set that will be approved based on predefined criteria. The characterisation data will be made available in machine-readable format. WP3 and WP7 will coordinate compound synthesis and develop new methods for characterisation of the CGL. WP1 will also establish an independent review mechanism governing CGL quality.
WP2 will: (i) Evaluate compound structural integrity and physiochemical properties, (ii) evaluate cellular potency of each compound against the primary target(s), (iii) evaluate compound selectivity against its protein family (if appropriate) and against the wider proteome, (iv) make all data available through the EUbOPEN web-based gateway (WP10) and through publication of comprehensive data packages.
WP3 will expedite the generation and characterisation of chemogenomics (CG) compounds by establishing novel and broadly applicable methods and technologies for biochemical, biophysical and cell-based assays, focused on multiplexed assay systems and multi-omics approaches. WP3 will also provide the ~2000-3000 additional compounds needed to complete the coverage of one third of the druggable genome (~1000 targets). WP3 will achieve this in part by sourcing externally generated high-quality CG compounds through a strong network of collaborations.
WP4 will generate protein expression clones and produce proteins and antibodies.
WP5 aims to (i) develop robust biochemical and biophysical assays suitable for hit discovery, validation, and optimisation, and family-wide assessment of chemical probe selectivity, (ii) develop cellular assays to confirm target engagement in cells, and (iii) generate CRISPR/Cas knockout cell lines for all targets to use as controls for validation of chemical probe activity and phenotypes in cells, as well as for characterising recombinant antibodies (WP4).
WP6 will (i) Solve 3D protein structures of targets with a relevant ligand (peptide, small molecule, substrate, etc), if not already known, (ii) Establish crystallisation conditions for fragment screening for E3 ligases, (iii) Determine high resolution structures of protein-ligand complexes to support structure-guided design of chemical probes and chemogenomic compound sets. Chemical starting points will be sourced from EFPIA partners, academic collaborators, DNA-encoded libraries and EUbOPEN.
The main objective of WP7 is to deliver 100 high quality chemical probes to decipher the biology of their annotated targets in phenotypic assays both within (WP9) and outside the consortium. The chemical probes and suitable analogues discovered over the course of the project will be added to the chemogenomics library (WP1).
The aim of WP8 is the development of chemical probes for significant parts of the human proteome is currently hindered by high cost and limited expertise. Beyond the immediate deliverables of this call, it will be essential to provide the larger scientific community with tools and access to platforms, which accelerate transformation of ideas into experiments and eventually into new probes. There are three major aspects of this call that require significant technology development: (i) Hit-to-lead chemistry. Converting initial chemical matter (e.g. fragments) into a potent lead series (hit-to-lead) is time- and cost-intensive by traditional medicinal chemistry methods. (ii) Compound selectivity. Current selectivity screens probe only a fraction of the proteome. It is essential to develop technologies to assess proteome-wide selectivity, (iii) Maximising use of patient samples. Studies using patient samples are limited by small quantities and heterogeneity. In order to address these limitations, we will develop methods to (i) Combine a web-based compound design platform with miniaturised, automated chemistry capable of accessing large and novel chemical space. (ii) Profile compounds in cells allowing the assessment of selectivity against the entire proteome, using a range of -omics approaches. (iii) Miniaturise and improve data quality from cellular and patient-tissue-based assays and develop reconstituted systems that will closely represent drug effects in vivo. Methods will be developed within EUbOPEN and by collaboration with leading laboratories. Importantly, all methods will be standardised and made available to the research community through collaboration or through detailed protocols. Moreover, we will achieve hardening of our methods by giving external users access to these platforms, in order to discover design flaws as soon as possible.
The goals for WP9 are (i) to characterise primary patient material and patient-derived renewable resources by multi-omics analysis, (ii) to develop and validate at least 20 new patient cell assays for irritable bowl disease (IBD) and colorectal cancer, (iii) to profile CGL compounds and chemical probes across the IBD and colorectal cancer patient cell assays, and also in other disease areas through partnerships, (iv) to develop complex co-culture systems to integrate different pathophysiological aspects of IBD and colorectal cancer (WP8), and (v) to publish the results and maximize the translational benefits from new discoveries.
WP10 will (i) establish compound logistics for efficient distribution of CGLs and chemical probes as well as compound exchange between partners, (ii) establish more than 500 assays for the annotation and profiling of CGLs and chemical probes, as well as relevant compounds from the community, (iii) build a database suitable for chemists and biologists that adheres to FAIR principles, (iv) establish long-lived platforms and transferrable infrastructure, and (v) establish an open access and dissemination framework.
WP11 will: (i) establish a framework for a global network of partners around the world that work on related goals, with a governance structure that supports efficient collaboration and sustainability, (ii) generate partnership agreements with major European and international efforts in related areas, and (iii) establish a process for recruitment and rigorous triage of external activity and contributions.
WP12 will provide a project management and governance structure to ensure the project meets its aims.



