Chemogenomics describes a method that utilizes well annotated and characterized tool compounds for the functional annotation of proteins in complex cellular systems and the discovery and validation of targets. In contrast to chemical probes , chemical small molecule modulators such as agonists or antagonist used in chemogenomic studies may not be exclusively selective. As high quality chemical probes have only been developed for a very small fraction of targets, the less stringent criteria for defining small molecules used in chemogenomics compound sets enables covering a larger target space. It is one of the main goals of EUbOPEN to cover about 30% of all targets that are currently thought to be druggable. To date, the druggable proteome has been estimated to comprise approximately 3000 targets. However, continued efforts exploiting new target areas such as the ubiquitin system and solute carriers which are focus areas in EUbOPEN, the number of proteins deemed to be druggable will continuously increase.
We defined within EUbOPEN criteria for inclusion of small molecules into the EUbOPEN chemogenomic library. These criteria have been peer reviewed by a committee of independent experts and are summarized here. Different scenarios covering a target space with a chemogenomic compound collection are depicted in the figure below.
Chemogenomic compound set
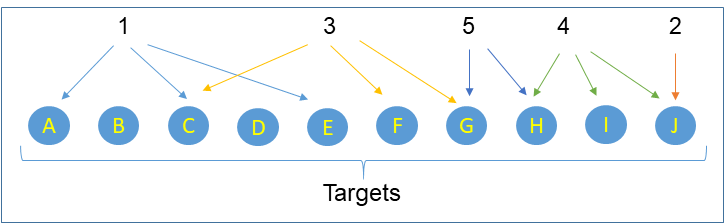
The chemogenomic set will be organized into subsets covering major target families such as protein kinases, membrane proteins and epigenetic modulators. The current chemogenomic target list is available here.



