Disease area
Colorectal cancer (CRC) is a highly heterogeneous disease and remains a leading cause of cancer-related mortality worldwide. Despite advancements in targeted therapies and immunotherapy, treatment resistance and tumor recurrence continue to be major clinical challenges. The complex molecular landscape of CRC, including genetic and epigenetic alterations, drives tumor progression and limits the effectiveness of current treatments. Novel therapeutic strategies targeting key oncogenic pathways are essential to improving treatment outcomes and overcoming resistance mechanisms in CRC.
Rationale
Patient-derived organoids (PDOs) are powerful preclinical models for drug screening, preserving the genetic and functional traits of the original tumor. Screening large pharmacological libraries in PDOs enables the systematic identification of novel CRC vulnerabilities and underlying mechanisms of action. PDO-based drug screening can help to develop of more effective therapies and to overcome therapy resistances. Further improvement of the available protocols for high-throughput screening will accelerate strategies for personalized medicine.
Aim
This study aims to optimize a high-throughput 384-well screening platform in CRC PDOs to measure drug effects at varying concentrations. Specific aims are to improve cell seeding and drug delivery. By integrating multiparametric readouts including cell viability and PDO morphology, this will enable a comprehensive analysis of drug responses, capturing a wide range of therapeutic effects.
Methods
Cell culture condition: CRC organoid line O12, which was used for assay development was established and maintained according to detailed protocols available on the EUbOPEN website (refer to Protocol for Generation of Normal and Tumor Organoids from Human Colorectal Tissues) and the recent publication by Farin et al., (2023).
General protocol: Organoids were cultured in 384-well, F-bottom plates (Greiner), which were pre-coated with 5 µL of 10% Basement Membrane Extract (BME) using a microfluidic liquid dispenser (Mantis, Formulatrix). Single-cell suspensions of organoids were prepared and seeded in 45 µL of complete organoid medium containing 10% BME. The organoid suspension was applied using a multichannel pipette, resulting in a uniform layer of organoids over the BME coating. On day 3, 20 µL of Calcein AM (1:1000, BioLegend) was added to each well to enable live-cell fluorescence imaging. Calcein AM stains viable cells, providing a measure of organoid viability. Automated imaging of the entire plate was then performed using a BioTek Cytation C10 imaging system (Agilent). Images were captured to assess organoid growth, morphology, and health. Data analysis was carried out using Gen5 software, to quantify organoid size.
Readout: On day 9, another 20 µL of Calcein AM (1:1000, BioLegend) was added to each well for live-cell staining, and the plate was imaged automatically to assess organoid viability. After imaging, 50 µL of medium was removed from each well using the automatic multichannel pipette (Integra), and 40 µL of CellTiter-Glo reagent was added. The contents of each well were mixed thoroughly to ensure proper reagent distribution. Luminescence was measured after a 30-minute incubation period to assess cell viability and metabolic activity, providing a quantitative readout of organoid health.
Results
The experimental workflow combines automatic cell seeding, automatic cell imaging, drug treatment, and enzymatic viability analysis, as illustrated in the schematic (Fig. 1A). Organoids were seeded on a BME layer in a 384-well format, providing support and ensuring image stability. To normalize well-to-well fluctuations, we used Calcein AM, a cell-permeable dye that undergoes hydrolysis by intracellular esterases, converting into a green-fluorescent signal. Staining was performed both before and after drug treatment on day 3 and day 9 (Fig. 1B), enabling baseline cell viability assessment and organoid uniformity evaluation across the plate. Organoid density was measured by sum areas in both the green channel (indicating viable cell area) and the brightfield channel (representing total organoid area). Additionally, CellTiter-Glo (CTG) measurement at the endpoint further supported the assessment of overall cell viability and metabolic activity across the plate (Fig. 1C-E). The coefficient of variation (CV) for organoid uniformity, measured without drug treatment, remained consistently low, ensuring high reproducibility and minimal well-to-well fluctuations in the assay.
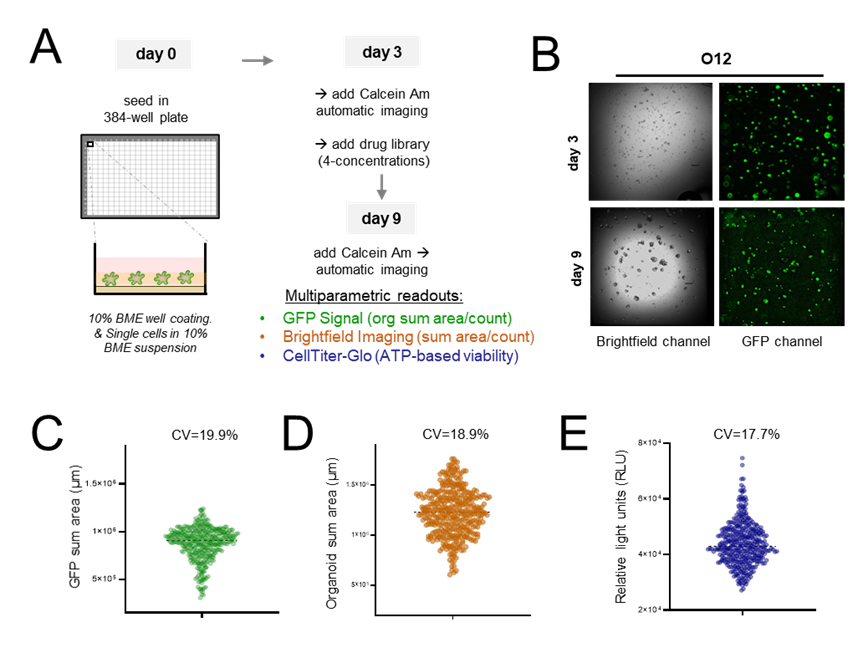
Figure 1. 384 well assay development of CRC organoids using multiparametric readouts. (A) Schematic representation of the screening workflow. Organoids were seeded in a 384-well format as single cells (day 0) on BME-coated wells and cultured in suspension medium containing 10% BME. On day 3, viability was assessed using Calcein AM staining, followed by automated imaging. On day 9, organoids were re-stained with Calcein AM and imaged to perform final viability measurements. (B) Representative brightfield and GFP images illustrate organoid morphology and fluorescent signal at day 3 and 9. (C-E) Multiparametric readouts included GFP signal (green; the sum of organoid area/count); brightfield imaging (orange; sum of organoid area/count), and CellTiter-Glo (blue, endpoint luminescence assay). Violin plots show the distribution of (C) GFP channel, (D) brightfield channel and (E) CTG readouts indicating a good seeding uniformity.
Conclusions
In conclusion, we present a robust and personalized preliminary platform for screening drug library. This platform integrates multiple imaging modalities, including brightfield, GFP signal, and endpoint luminescence (CellTiter-Glo), to assess organoid morphology, growth, and metabolic activity. By leveraging these multiparametric readouts in a high-throughput format, our goal is to screen a comprehensive drug library in a concentration-dependent manner, identifying novel drug vulnerabilities.
References
- Farin HF, Mosa MH, Ndreshkjana B, et al. Colorectal Cancer Organoid-Stroma Biobank Allows Subtype-Specific Assessment of Individualized Therapy Responses. Cancer Discov. 2023;13(10):2192-2211. doi:10.1158/2159-8290.CD-23- 0050



