Disease area
Colorectal cancer (CRC) remains one of the most lethal malignancies and is often diagnosed at a progressed stage when tumor cell dissemination has already started. The tumor microenvironment (TME) plays a crucial role in regulating malignant tumor characteristics including growth, metastasis, and therapy resistance. Cancer-associated fibroblasts (CAFs) are one of the main cell types in the TME that show considerable heterogeneity and plasticity. Through different processes, such as paracrine signalling or metabolic changes, CAFs can modulate the cancer phenotype.
Rationale
CAFs can modulate the efficiency of organoid colony formation in vitro and/or promote the subsequent tumor cell growth. In a reciprocal fashion, organoids can influence the growth and polarization of CAFs. Here we want to identify pharmacological compounds that interfere with the crosstalk between tumor and stromal cells using a compartment-specific viability screening approach.
Aim
We have developed a dual luciferase assay to monitor the sensitivity of chemogenomic libraries in a compartment-specific manner in CRC organoid-stroma co-cultures.
Methods
Cell culture: PDTOs were established and cultured as previously described (van de Wetering et al., 2015). Tumor cells are cultured in full medium containing advanced DMEM/F12 supplemented with 10 mM Hepes, 1× Glutamax, 1× penicillin/streptomycin, 2% B27, 12.5 mM N-acetylcysteine, 500 nM A83-01, 10 μM SB202190, 20% R-spondin 1 conditioned medium, 10% Noggin conditioned medium, 50 ng/ml human EGF. Fibroblasts were cultured in growing medium containing DMEM, 10% FCS and 1% penicillin/streptomycin.
General protocol: To measure compound effects in co-culture, organoids were transduced with Luciferase2-P2A-EGFP lentivirus as described (Schnalzger et al., 2019) and fibroblasts were transduced with Renilla-dsRed lentivirus. The lentiviral constructs are available upon request. We screened co-cultures of O06 + F06, O14 + F14 and O20 + F20. Single organoid cells were seeded in presence of CAFs in 96-well plates in 15 µl 70% Matrigel and grown for 3 days in full organoid medium (100 µl/well). On day 3, cells were washed and cultured with growth factor-reduced medium in presence of chemogenomic library drug for 6 days. The chemical probe and chemogenomic library used in this screen contained 262 active and 108 negative control compounds. The screen was conducted in 2 independent replicates.
Readout: Dual-Glo Luciferase assay (Promega) was used as readout on day 9.
Results
To identify compartment-specific responses in organoid-stroma co-cultures, we developed the Dual-Glo luciferase system where we first measure the firefly luciferase signals generated by organoids. Then Stop&Glo reagent quenches the firefly signal followed by measurement of renilla luciferase signals from fibroblasts (Fig. 1A). Transduced Luciferase-GFP organoids and Renilla-dsRed fibroblasts are captured by confocal imaging with fibroblasts (red) in close proximity to the organoids (green) (Fig. 1B). Strong cytotoxic effect after Staurosporine treatment observed in both compartments (Fig. 1C). Pharmacological treatment in co-culture exposed compartment-specific drug sensitivity in three patient-derived models (Fig. 1D-E). Comparison of top hits among the 3 models (Venn diagrams) identified shared targets but also patient-specific vulnerabilities in tumor cells (Fig. 1G), and in CAFs (Fig. 1H).
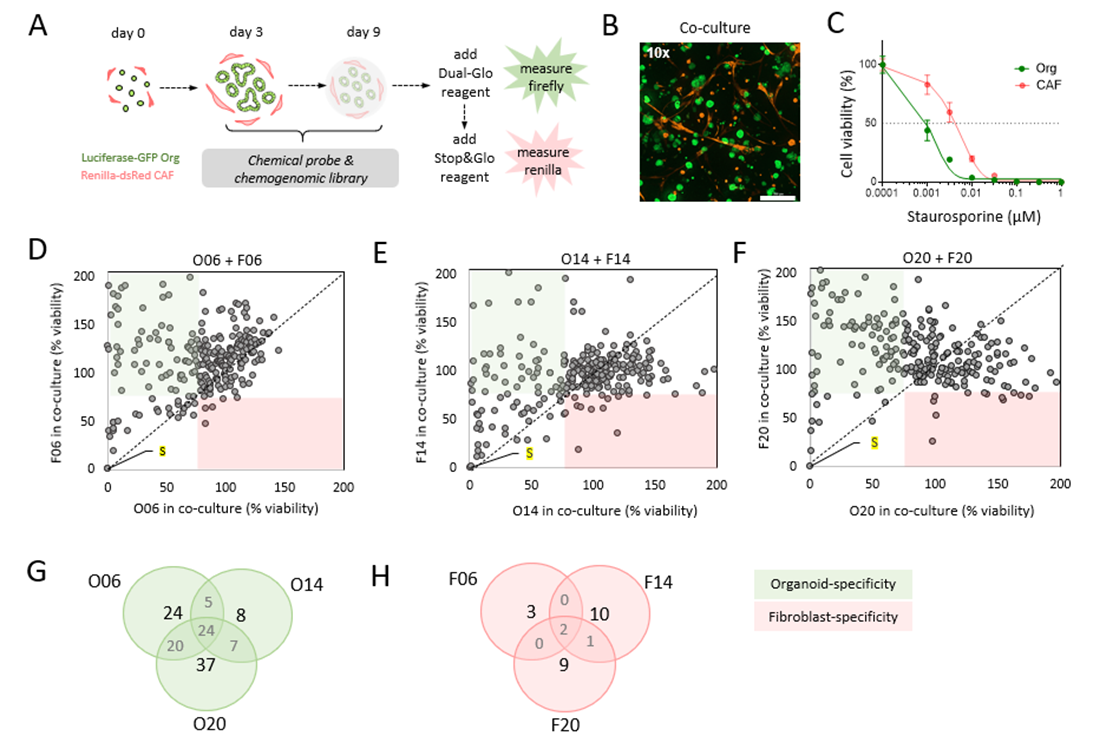
Figure 1. Identification of compartment-specific drug sensitivities in CRC organoid-stroma co-cultures. A) Schematic design of Dual-Glo luciferase assay to assess drug library sensitivities in co-cultures. B) Co-culture of CRC organoids and fibroblasts that were transduced to stably express firefly luciferase-GFP and renilla luciferase-DsRed, respectively. Confocal image stacks of GFP and DsRed is shown at (10×) magnification. Scale bar is 300 μm. C) Cell viability measurement of Staurosporine treatment in co-culture. Strong cytotoxic effects are shown in both compartments. D-E) The chemical probes and chemogenomic library containing 370 compounds (including controls) was screened in O06, O14 and O20 organoids co-cultured with their autologous fibroblasts. Compartment specific vulnerabilities are measured after 6 days of treatment and cell viability was assessed by Dual-Glo measurement. Green quadrants depict organoid-specific sensitivity, where the red quadrants show the fibroblast-specific vulnerabilities. Cut-off of 75% viability was used to identify compounds that differently target stromal or tumor cells. Mean data from 2 experimental replicates is shown (negative controls are left out from the plots). Staurosporine (S) treatment is labelled in yellow. G-H) Venn diagram shows inter-patient differences. G) Cut-off of ≤ 75% viability in organoids and ≥ 75% viability in fibroblasts was used to identify compounds that disturb only tumor organoid growth. H) Similar analysis was performed for identification of stroma-specific targeting (cut-off of ≤ 75% viability in fibroblast and ≥ 75% viability in organoids).
Conclusions
Together, our results show that dual reporter co-cultures can be used to screen pharmacological libraries to identify drugs that selectively affect tumor cells or CAFs providing a proof-of-principle for translation of the chemogenomic libraries.
References
- van de Wetering et al., 2015, Cell 161, 933–945.
- Schnalzger et al., 2019, EMBO 238(12): e100928



