Disease area
Colorectal cancer (CRC) is a major contributor to cancer-related deaths globally that is often detected at advanced stages when tumor cells have already disseminated. Despite progress in chemotherapy and targeted treatments, therapy resistance remains a significant challenge in clinical practice. Within the tumor microenvironment (TME), cancer-associated fibroblasts (CAFs) have emerged as a critical component, influencing tumor progression and therapy response.
Rationale
Understanding the tumor microenvironment is vital for effective cancer therapy. To accurately study tumor-stromal interactions, scalable experimental models are required that integrate CAFs. By dissecting how CAFs influence tumor behavior we can identify actionable targets and develop more effective treatments.
Aim
Building upon our prior work of a CRC organoid-stroma biobank (Farin et al., 2023), we have now developed an Optimized DualGlow Luciferase Assay to simultaneously monitor both compartments (tumor cells and CAFs) in co-cultures. A destabilized Renilla construct was used to reduce the enzymatic half-life and allow more accurate assessment of cell viability after drug treatment. We use this assay to investigate the context-specific dynamics of tumor-stroma interactions after chemogenomic library screening in order to understand the intricate interplay between tumor and CAFs.
Methods
Cell culture: Organoids and matched CAFs were established and cultured as described in detail in the protocols deposited in the EUbOPEN website (Protocol for generation of normal and tumor organoids from human colorectal tissues & Protocol for generation and culture of CRC fibroblasts).
General protocol: To measure compound effects in co-culture, organoids were transduced with Luciferase2-P2A-EGFP (Luc2/GFP) lentivirus as described (Schnalzger et al., 2019) and fibroblasts were transduced with the original Renilla-dsRed lentivirus (Ren/DsRed; Farin et al., 2023), a codon optimized Renilla-dsRed (Ren (co. opt.)/DsRed), and a codon optimized Renilla-dsRed (Ren*/DsRed) lentivirus, which additionally includes two protein destabilization sequences; hCL1 and hPEST fragments (E7521, Promega) (Fig. 1A). Matched single organoid and CAF cells from three patients #06, #14 and #20 (Farin et al., 2023) were seeded in mono- and-co-cultures in 96-well plates in 15 µl 70% Matrigel and grown for 3 days in full organoid medium (100 µl/well). On day 3, cells were washed and cultured with growth factor-reduced medium in presence of the DCP library incl. chemogenomic kinase set (370 compounds incl. the negative control compounds). The screen was conducted in 2 independent replicates.
Readout: Dual-Glo Luciferase Assay System (E2920, Promega) was used as readout on day 9, following the manufacturer’s instructions.
Results
To enhance the sensitivity of the Renilla luciferase reporter, we refined our previous construct by introducing a codon-optimized Renilla luciferase gene along with two protein destabilization sequences (Fig. 1A). Codon optimization significantly boosts luciferase expression levels. The incorporation of the two destabilization sequences reduces the enzymatic activity, but permits a more accurate measurement due to the reduced enzymatic half-life (Fig. 1B). Notably, upon treatment with Staurosporine, we observed a reduced variability and a dose-response curve that mirrored the Luc2 read-out that was measured in parallel (Fig. 1C).
For monitoring compound effects in co-culture settings, we devised an assay wherein both cell compartments were marked (Fig. 2A). Screening was conducted in matched co-cultures from n=3 patients to discern response patterns. Pearson correlation analysis and hierarchical clustering revealed similarity of experimental replicates and distinct responses in CAFs and organoids (Fig. 2B). To identify CAF induced resistance or vulnerability, we examined the response of organoids in both mono- and co-cultures. Intriguingly, inhibition of certain targets rendered organoids either more resistant or more sensitive. CAF-induced signaling appears to compensate for inhibition of the MAPK-RAS signaling pathway, while FGFR1 inhibition makes organoid cells more vulnerable in co-culture. Analysis of the top hits from 3 patients that rendered the organoids more vulnerable showed two shared targets, BLC2 and FASN, in addition to numerous patient-specific hits (Fig. 2C). In parallel, we observed that the presence of organoids also protects CAFs or renders them more sensitive to treatment (Fig. 2D).
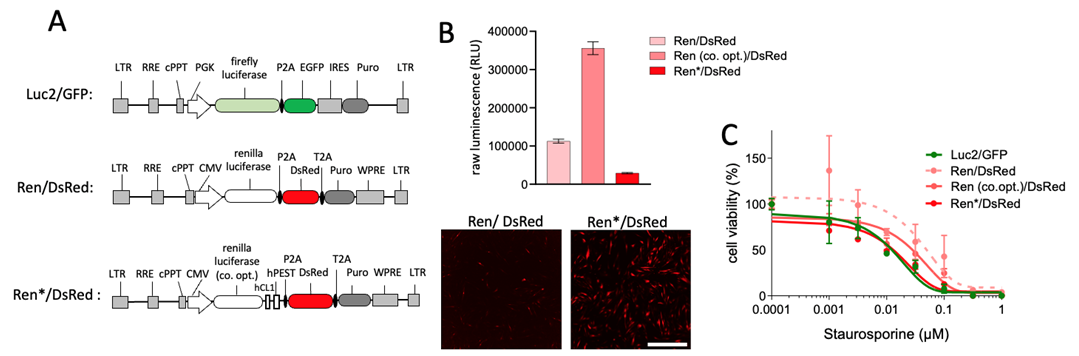
Figure 1: Optimization of renilla luciferase lentivirus in CAFs. A) Schematic representation of the optimized construct integrating codon-optimized renilla luciferase and two protein destabilization sequences Ren*/DsRed. B) Raw luminescence measurement of F14 fibroblast line transduced with the different reporters. The confocal image stacks of DsRed are shown at low (2.5×) magnification. Scale bar is: 2000 µm. C) Dose-response curve illustrating the enhanced sensitivity of Ren*/DsRed to Staurosporine treatment with the optimized construct.
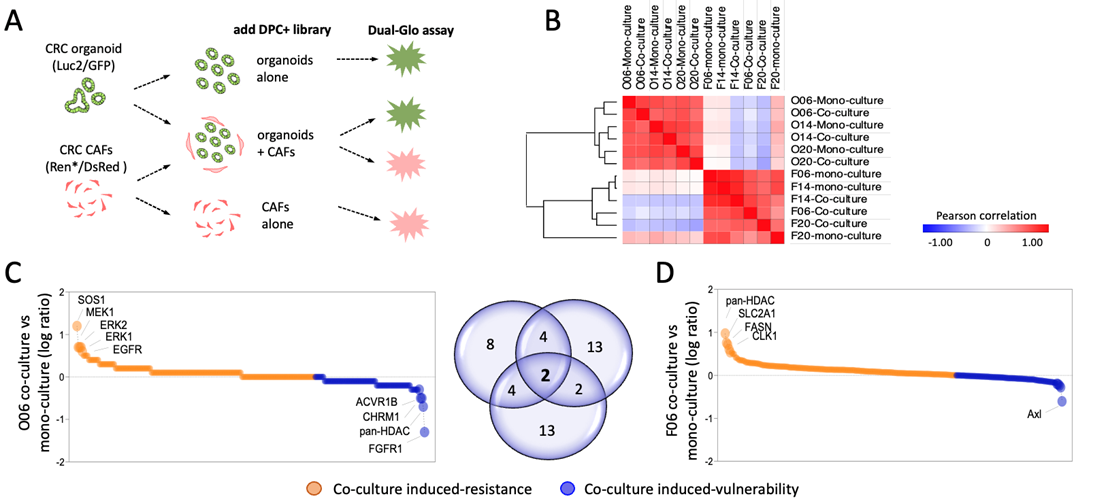
Figure 2. Perturbation of the tumor-stroma crosstalk. A) Assay design for monitoring compound effects in mono and co-cultures. B) Pearson correlation and hierarchical clustering shows distinct responses of organoids and CAFs following treatment with the DCP+ library. C) Relative drug response in organoids (O06). Orange denotes increased resistance and blue higher sensitivity in presence of CAFs. Key hits are labelled. D) Reciprocal analysis in CAFs shows a distinct response pattern with specific targets that are highlighted.
Conclusions
The described assay allows a reliable and scalable read-out to simultaneously examine tumor and CAF compartments in 3D CRC models. We found that both compartments mutually influence each other and identify that the co-culture can induce pharmacologic resistances and vulnerabilities. This platform represents a valuable tool for advancing our understanding of cancer biology and improving therapeutic strategies targeting the tumor-stroma crosstalk.



