Disease area
Colorectal cancer (CRC) is among most lethal malignancies in the world and is often diagnosed at an advanced stage when the tumor cell dissemination has already started. Chemo- and targeted therapies provide only a limited increase of overall survival for these patients. The major reason for clinical failure remains therapy resistance. New combination strategies that target cellular pathways that are rewired in tumor cells could help to overcome therapy resistance but the identification of actionable drivers for personalized therapy remains challenging.
Rationale
Patient-derived tumor organoids have recently emerged as preclinical models that faithfully recapitulate the molecular and phenotypic characteristics of CRC. We have established a CRC organoid biobank and have molecularly characterized 29 patientderived organoids (PDOs). To model the response to chemotherapy, we have exposed all tumor organoids to 5-FU, Oxaliplatin, SN-38 and Gefitinib and measured individual sensitivities. This allowed us to identify resistant tumor samples providing an opportunity to study underlying mechanisms.
Aim
In order identify strategies for therapy re-sensitization we will subject resistant CRC organoids to sub-toxic doses of therapeutic drugs in combination with chemical probe libraries.
Methods
Cell culture condition: PDOs were established and cultured as previously described (van de Wetering et al., 2015). For detailed information on organoid handling and culturing refer SOP. Tumor cells are cultured for three days in 50 µl/well medium containing advanced DMEM/F12 supplemented with 10 mM Hepes, 1× Glutamax, 1× penicillin/streptomycin, 2% B27, 12.5 mM Nacetylcysteine, 500 nM A83-01, 10 μM SB202190, 20% R-spondin 1 conditioned medium, 10% Noggin conditioned medium, 50 ng/ml human EGF.
General protocol: Colony formation assay was performed in n=29 CRC organoids to standardize the input cell number. Organoids were transduced with Luciferase2-P2A-EGFP lentivirus as described (Schnalzger et al., 2019). In a 96 well format, single cells were seeded in 15 µl 90% Matrigel, grown for 3 days in full organoid medium. On day 3, cells were washed and cultured in growth factor-reduced medium in presence of therapeutic drug for 6 days. Sensitivity to 5-FU, Oxaliplatin, SN-38 or Gefitinib was tested at 7-point dilution using a digital dispenser (Tecan De300). Combination screens of tumor organoids will be performed in 384-well plates. Cells will be enzymatically dissociated, seeded in 10 µl 50% Matrigel and let recover for 3 days before culture in growth factor reduced medium. 5-FU, Oxaliplatin, SN-38 or Gefitinib will be added at fixed a sub-toxic concentration (determined above) and combined with chemical probe library that will be screened at a 4-point dilution for 4 days.
Readout: Luciferin Live Cell Substrate will be performed as readout on day 3 and the viability was measured after 7 days of culture by One-Glo EX Luciferase assay system. To correct for seeding differences the data will be normalized in each well to the initial measurement.
Results
Colony formation assay in 29 lines showed different growth potential of single cells. Therefore, we standardized the input cell number to assure similar colony number for all lines (Fig. 1A). Treatment of organoids with the chemotherapeutic drugs 5-FU, Oxaliplatin, SN-38 and the EGFR inhibitor Gefitinib, revealed a heterogenous response. Drug sensitivity is plotted as normalized Area Under the Curve (AUC) (Fig. 1B).
The resistant lines for each individual drugs (marked in red) will be subjected to chemical probe libraries to identify chemotherapy sensitizers. In 384-well plates, organoid cell will be subjected to either compound libraries alone or in combination with a sublethal dose of therapeutic drug. We will the identify chemical probes that only show toxicity only in combination combined (Fig. 1C). In preparation of the screens, seeding consistency in 384-well plates was confirmed (Fig. 1D). For quality control (Z-factor and Z-Prime factor) each screening plate will contain positive and negative controls. Viability data after single agent and combination treatment will be fitted analysed to determine IC50, AUC and Drug Scoring Sensitivity (DSS). To score for synergistic combinations, we will use Bliss independence model that compares observed and the predicted combinatorial responses.
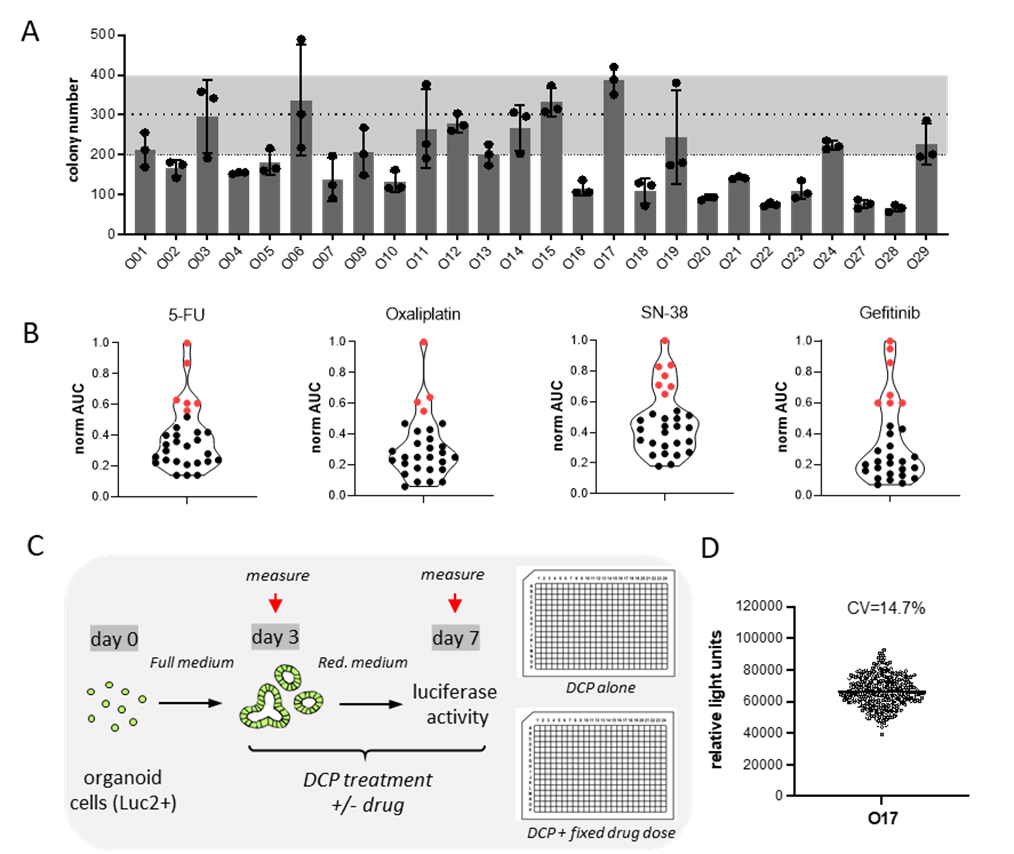
Figure 1. Therapy re-sensitization of drug resistant organoids. (A) Colony formation assay for 29 CRC organoids. (B) Sensitivity to different chemotherapies and the targeted compounds (Gefitinib). Resistant lines are marked in red. (C) Schematic representation of experimental design of therapy re-sensitization assay. (D) Seeding consistency of untreated 384-well plate. Dot plot of raw data (relative light units). Coefficient of variation (CV) is shown.
Conclusions
We present a powerful personalized platform to screen chemogenomic drug libraries to overcome therapy resistance. Screening data will be updated as available.
References
- van de Wetering et al., 2015, Cell 161, 933–945.
- Schnalzger et al., 2019, EMBO 238(12): e100928



